Libra – Structural Logic 🧠
The Challenge of Temporal Reasoning in Radiology AI
When dealing with radiology images, especially in the context of temporal analysis—comparing current chest X-rays with previous images—standard neural network architectures often struggle. Although transformer-based multimodal large language models (MLLMs) like LLaVA demonstrate remarkable capabilities for understanding single images and textual information, they encounter substantial challenges when handling image pairs.
However, capturing meaningful temporal differences between two images remains problematic with traditional transformer structures.
When Transformers Lose the Plot: Why They Struggle with Temporal
The transformer, the cornerstone of modern large language models (LLMs), excels at sequential data processing and logical reasoning tasks. Its strength lies in handling complex linguistic structures through positional encoding, enabling nuanced relationships in textual sequences.
However, when transformers receive visual information—particularly multiple images presented simultaneously—the situation becomes more complicated. Existing methods typically concatenate image features directly into the LLM’s head, often via sequences containing hundreds of visual tokens (patch tokens), depending on the specific image encoder used.
Indeed, current MLLMs like LLaVA perform impressively with single-image inputs. But they quickly become overwhelmed with paired images, heavily relying on meticulously crafted instruction datasets to guide temporal comparisons explicitly:
How MLLMs Are Prompted to Compare Images
“What is the difference between <image-1-patchholder> and <image-2-patchholder>?”
Such approaches place the burden squarely on the LLM’s internal reasoning and positional encodings, complicating training and diminishing reliability. The model must:
- Distinguish between multiple images using only position encodings
- Process 500+ tokens per image (depending on patch number)
- Compare features across long token distances
Given these limitations, an essential question arises:
Can we overcome these temporal reasoning challenges structurally, rather than through explicit prompting?
Structure Determines Function: Insights from Biology
Before we answer above question, let’s briefly reflect on the foundational relationship between structure and function—deeply ingrained in biological systems.
Macro-scale examples:
- Birds have wings enabling flight
- Fish possess gills allowing them to breathe underwater
Micro-scale examples:
- The unique three-dimensional helical structure of proteins directly determines their biological roles
- A virus’s outer shell dictates its infection pathways and interaction mechanisms
Clearly, function is fundamentally dependent on structure.
When designing novel neural architectures or modules, we must apply this principle:
- Identify the desired functionality first
- Then craft an appropriate structural design that inherently supports these functions
Libra’s Structural Innovation
Temporal Alignment Connector (TAC)
Following this logic, we developed the TAC in our Libra model. TAC’s primary goal is to automatically and effectively capture the relationship between two chest X-ray images—the current image (primary) and a prior image (auxiliary).
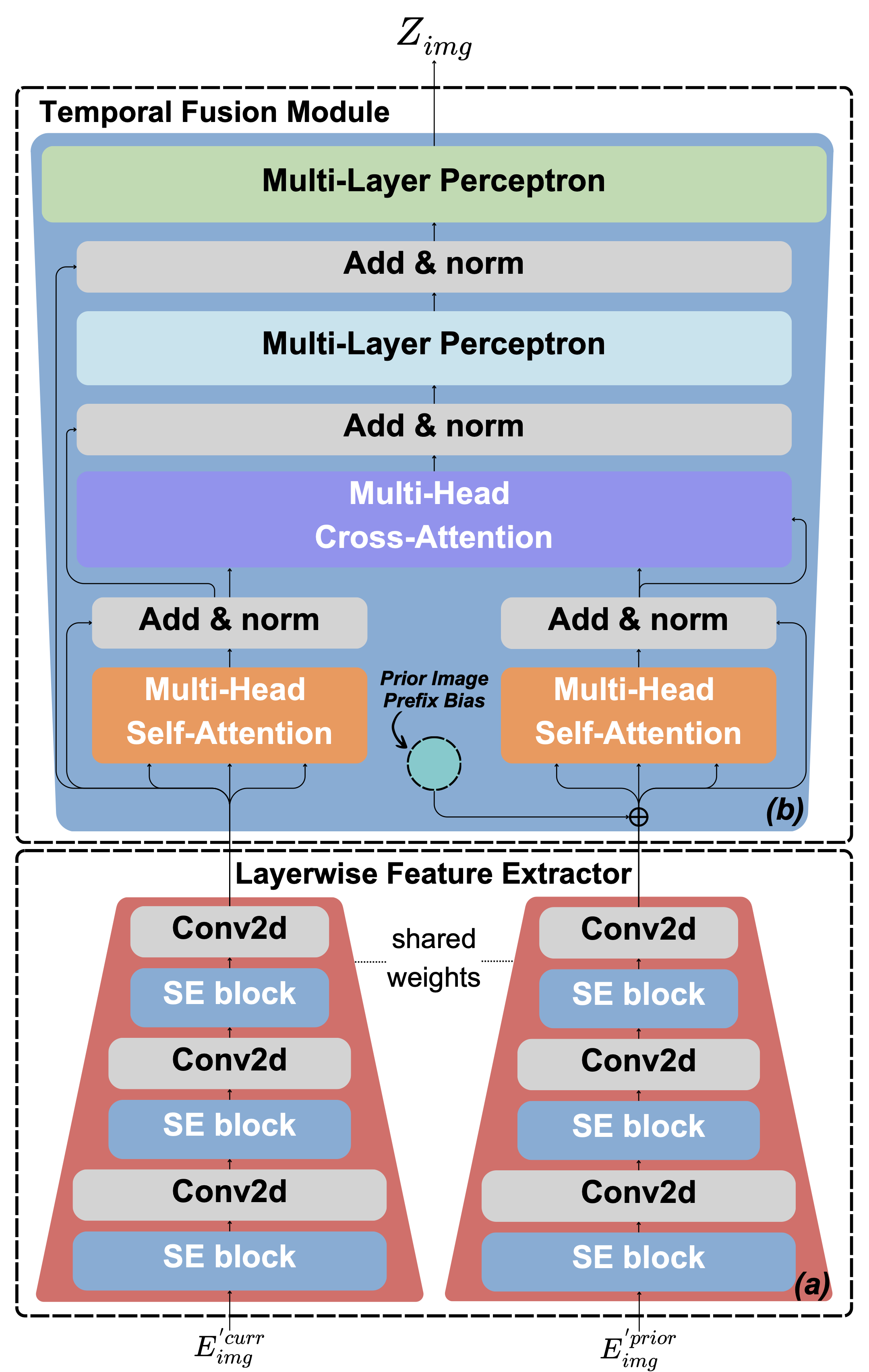
Libra’s Temporal Alignment Connector (TAC) architecture.
Unlike traditional transformers that treat all inputs equivalently, TAC explicitly structures interactions between paired images. It captures their nuanced relationship through two key modules:
- TAC Architecture
- Layerwise Feature Extractor (LFE)
- Aggregates visual features across multiple encoder layers
- Ensures rich representations from both images
- Maintains feature hierarchy information
- Temporal Fusion Module (TFM)
- Fuses features from current and prior images
- Highlights critical temporal differences
- Maintains clear image role assignment
- Current image (Primary)
- Prior image (Reference)
- Prefix Bias Mechanism
- Addresses nearly-identical image pairs
- Prevents attention collapse
- Differentiates prior image's contextual influenceAn important structural consideration is the integration of a prefix bias mechanism. This component addresses the scenario where current and prior images are nearly identical—common in clinical practice. Without careful design, such similarity can cause attention mechanisms to collapse into redundant self-attention loops.
Why Structure Matters: The Libra Advantage
By structurally encoding temporal relationships directly into the neural network’s architecture, Libra overcomes the limitations inherent in traditional prompting-based approaches. Instead of forcing the LLM to implicitly infer temporal differences through complex positional encodings and exhaustive instruction tuning, TAC explicitly and efficiently captures this essential clinical context.
Libra exemplifies the powerful concept that structural logic, thoughtfully aligned with functional requirements, dramatically enhances model performance.
This structural logic not only simplifies training but also improves:
- Reliability: More consistent temporal reasoning
- Interpretability: Clearer connection between features and outputs
- Efficiency: Reduced dependence on instruction tuning
- Clinical Alignment: Better reflection of radiologists’ actual workflow
🏄 Note: The opinions shared here reflect my own understanding and are intended to convey the structural logic behind Libra. For technical accuracy and complete details, please refer to our paper: “Libra: Leveraging Temporal Images for Biomedical Radiology Analysis”.